Equipe : Diversité, Evolution et génomique des Interactions Biotiques<br />
<br>
Téléphone : 02 23 48 51 31<br/>
Anne-Marie.Chevre@inrae.fr
Team leader: Biodiversity and Polyploidy
https://cv.archives-ouvertes.fr/anne-marie-chevre
Expertise-Skills
Polyploidy, recombination, interspecific hybridization, cytogenetics
Career
- Since 2007 - Team leader of the group Biodiversity and Polyploidy
- 2012-2016 - Deputy head of the new Research Unit, UMR1349 IGEPP INRA, AgroCampus Ouest- Université de Rennes1, Fr (more than 270 persons),
- 2009-2011 - Head of research unit, UMR118 APBV, INRA, AgroCampus Ouest- Université de Rennes1, Fr (more than 150 persons)
- Since 1995 - Senior scientist (at INRA, UMR APBV then IGEPP, Le Rheu, Fr.
- 1988-1989 - Post-doc at the Department of Vegetable Crops, University of California, Davis “Molecular characterization of B.napus-B.nigra addition lines”. Laboratoire de C.F.Quiros,
- 1985-1995 - Permanent position as scientist (CR) at INRA, Research Unit APBV, Le Rheu, France
- 1985 - PhD University of Bordeaux II (1985) Research on “In vitro vegetative micropropagation of chestnut”. Research Unit INRA Bordeaux (Dr G. Salesses)
Research interests
My research is mainly focused on Brassiceae species with complementary studies on the process of polyploidy stabilization through regulation of homologous and homoeologous recombination in Brassica. These knowledges are useful to insert genes of interest from related species into crops, and to assess gene flow from the crop to the weeds.
- Polyploidy stabilization through regulation of homologous and homoeologous recombination
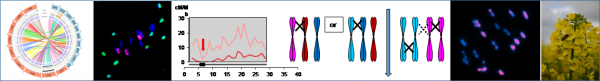
The Brassica model is particularly relevant to explore the process involved in polyploid stabilization and in recombination regulation. In fact, several rounds of whole genome duplication are at the origin of the different diploid Brassica species. A recent new round of polyploidy is at the origin of oilseed rape (Brassica napus, AACC, 2n=38) which is a natural hybrid between B. rapa (AA, 2n=20) and B. oleracea (CC, 2n=18). However, B. napus presents a very low genetic diversity due to high selection pressure for seed quality whereas a large diversity (see our Genetic Resource Center, BrACySol-/About-IGEPP/Platforms/BrACySol) exists in its diploid progenitors (e.g. Aissiou et al. 2018). In order to describe this huge diversity, we notably contributed to the sequencing of reference genomes of the diploid and polyploid species using the latest technologies (e.g. Belser et al. 2018).
We analyzed the different ways to introduce novel genetic diversity in B. napus using its diploid parental species. Depending on the chosen strategy, different impacts on either homologous or homoeologous recombination may be observed. From this research, we increase our knowledge on the genetic mechanism involved on polyploid stabilization (Pelé et al. 2018) and on possible genetic diversity enlargement.
First, we tried to clarify how diploid genome evolved in a polyploid context. To that purpose, we extracted by successive crosses the A genome of oilseed rape at the diploid AA stage and observed that it is impossible to obtain a AA diploid individual (2n=2x=20) with the A genome of B. napus, indicating an evolution in polyploid context (Pelé et al. 2017a). Secondly, we resynthesized oilseed rape by crosses between different varieties of the two diploid progenitors through different pathways. The first meiosis of the new oilseed rape acts as a genome blender promoting homoeologous recombination between A and C genomes (Szadkowski et al. 2010, 2011) as so generating non-reciprocal translocations in the following generations (Rousseau-Gueutin et al. 2017). The functional impact of gene redundancy and of structural rearrangements (homoeologous exchanges) on speciation are currently studied by Mathieu Rousseau-Gueutin. Sequencing data of this material already allowed elucidation of nucleo-cytoplasmic interactions in our polyploidy model (Ferreira de Carvalho et al. 2019). Additionally, we are analyzing the relation between genome dynamics and meiotic stability linked to seed fertility. Thirdly, we tested another strategy by direct crosses between oilseed rape and its progenitors and showed it is the most efficient technique to improve homologous recombination (Leflon et al. 2010).
All these data provided information on regulations of meiosis. In collaboration with E. Jenczewski (IJPB, Versailles, France), we showed that homoeologous recombination between A and C genome at the haploid stage is under polygenic control with a major QTL on C09, PrBn (Jenczewski et al. 2003; Liu et al. 2006; Nicolas et al. 2009, 2012; Grandont et al. 2014). The origin of this control, pre-existing in diploid or occurring in polyploidy context is still under debate. On another hand, we showed that it is possible to change drastically the regulation control of homologous recombination between A genomes in AAC allotriploid hybrid obtained by direct crosses between B. napus and B. rapa. The crossover frequency is increased (x3.4) and crossovers are distributed all along the A chromosomes even on genomic regions, which are normally deprived of crossover (pericentromeric regions) compared to the results obtained from diploid AA hybrids with the same A genotypes (Leflon et al. 2010; Pelé et al. 2017). This effect is not linked to the number of C chromosomes in addition but to specific C chromosomes, especially C09 (Suay et al. 2016; Pelé et al. in prep). From these data, the impact of the ploidy level on meiosis regulation and the identification of the mechanisms involved are in progress (PhD F. Boideau).
- Project France génomique PolySuccess (2014-2020) Coordination A.M. Chèvre
- Project BAP RecOptic (2019-2020) Coordination A.M. Chèvre
- Project Tassili (France-Algérie) (2016-2018) Coordination A.M. Chèvre et H. Hadj-Arab
- Project Promosol, Bingo (2016-2017) Coordination A.M. Chèvre
- Project ANR Blanc CROC (2014-2018) Coordination E. Jenczewski (IJPB, Versailles)
- Project INRA Transfert HyperRec (2013-2016) Coordination R.Mercier (IJPB, Versailles)
- Project ERA CAPS Evo-Genapus (2014-2016) Coordination I. Bancroft (York Univ., UK)
- Project BAP RecMax (2014-2015) Coordination A.M. Chèvre
- Project ANR Blanc CoPath (2007-2010) Coordination : C. Mézard (IJPB, Versailles)
- Project ANR Biodiversité (2006-2009) Co-Coordination avec M. Ainouche (Université Rennes 1)
- Project Barrande (France-République Tchèque) (2008-2009) coordination A.M. Chèvre-A. Kovarik
- Introgression of genes of interest within the crop
The knowledge acquired on regulation of recombination allowed introgression of gene of interest from related species in oilseed rape.
The high frequency of crossover between homologous chromosomes and the modification of recombination landscape in allotriploid hybrids leaded to a large research program initiated by S. Paillard (previously scientist in the group), aiming to enlarge oilseed rape diversity with introgressions of a limited size. Using a B. rapa and B.oleracea core collection, we performed crosses with the same B. napus variety, and produced large pre-breeding populations after several backcrosses with the same B. napus variety. The genotyping and the screening for gene of interest are in progress but seeds are already available and tested for various agronomic traits of interests by private partners. In parallel, the same strategy allowed introgression of a specific resistance gene to blackleg from B. rapa to B. napus (Balesdent et al. 2013).
Different strategies were developed from less closely-related species (B. nigra or B. juncea) to promote introgression into oilseed rape genome through homoeologous recombinations (Mason and Chèvre 2016) and to introduce specific blackleg resistance genes. Two of them, Rlm6 and Rlm10 genes will be soon available for private partners. Rlm6 was largely used to assess durability of specific resistance gene combined or not with a polygenic partial resistance (Brun et al. 2010)
- Project PIA Rapsodyn (2011-2020), Coordination N. Nesi (IGEPP)
- Project KBBE Gewidis (2014-2016) Coordination R.Delourme (IGEPP)
- Project Promosol, ProBiodiv (2011-2014) Coordination A.M. Chèvre
- Project ANR07 GPLA 07-024C AVirLep (2008-2011) Coordination: M. Balesdent (BIOGER)
- Project ADD-ANR-CEDRE (2006-2009) Co-Coordination avec L. Bousset (BIO3P, Rennes)
- Project UE FP5 SECURE (2002-2006) Coordination: N. Evans (Rothamsted, UK)
- Project UE FAIR (1998-2001) NORDIC Coordination : K. Glimelius (SLU, Uppsala, Sweden)
- Gene flow from oilseed rape to its weeds
The development of transgenic (herbicide tolerant) oilseed rape in 1988 leaded to the question of gene flow from the crop to its weeds. After testing several weeds for their probability to produce interspecific hybrids (hand pollination and field experiments), we found that wild radish (Raphanus raphanistrum, RrRr, 2n=18) was one of the weed able to produce number of interspecific hybrids. After five generations under field conditions with pollination of hybrids with wild radish, we observed that three B. napus genomic regions had the highest probability to be introduced into wild radish genome, indicating that gene flow depends on initial location of the transgene into the crop (Adamczyk-Chauvat et al. 2017). We are trying to introduce bar gene in hot and cold reions using CRISPR-Cas9 technology in order to validate these results with herbicide selection pressure on larger populations under confined greenhouse conditions
- Project PIA Genius (2012-2020) Coordination P. Rogowsky (ENS Lyon)
- Project ANR OGM Natora (2008-2011) Coordination A.M. Chèvre
- Project ANR MFD/AO OGM 0217 (2007-2009). Coordination B. Hau (CIRAD)
- Project ANR-06-POGM02 COBINA (2007-2010) Coordination : P.B. Joly (Evry)
- Project ACI OGM : (2002-2005) Co-Coordination avec E. Jenczewski (IJPB, Versailles)
- Project Ministère de l’Environnement Etude des barrières interspécifiques (1995-1996), Coordination A.M. Chèvre
Research animation
- Member of Editorial boarding of Plant Breeding since 2001, BMC Genomics since 2016
- Expert of national and international projects (BAP, RFI Végétal, BRG, ACI, ANR, MENRT, CTPS, ANR, ERC, Austian Science Fund, NWO Wageningen)
- Member of INRA national scientific Committee GAP until 2011 and BAP since 2016
- Member of several scientific committees: Risk in Ecology Ministry (since 2011), ANR Systerra (2009), Research Station UPR Montpellier 2005, UMR AGAP Montpellier (2017), Member of AERES commission 2010, Evaluation of CIRAD Scientists (2010)
- Members of scientific committees of international congresses: ESF Brassica gene flow (2001), International Conference on Polyploidy, Hybridization and Biodiversity ICPHB (2009), Crucifer workshop Catane (2012)
- Members of scientific committees for different socio-economic partners: President of Vegenov COST since 2013, member of Vegepolys COST since 2013
Teaching and Formation
- Supervisor Master2 and PhD
- Examiner of grant competition, reporter thesis committees and HDR
- Member of the council of doctoral school EGAAL since 2017
- Leader for the animation of PhD group (~70PhD/year) in Plant Breeding department (1997-2013).
- Animation of public debates on GMO, polyploidy or durability
- Interviews by journalists